Start your query
Click the tab 'Query' on the navigation bar at the top of the website.
This will display the empty form for submitting your query:
Submit your query
Insert your protein sequences in the input field.
This can be done in the following ways:
- Directly paste the sequences into the input field.
- Upload the sequences from a file by clicking 'Upload from file'.
- Use the example provided by the webpage by clicking 'Paste example'.
After inserting the sequences in the input field, submit the query by clicking the 'Submit'-button.
After submitting your query
If the submit was successful, your query-webpage will look like this:
Please copy the displayed link from the flash-message. Afterwards, you do not need to keep the page open and can use the link to return to your query.
Get results from your query
As soon as your query finishes, your query-webpage will look like this:
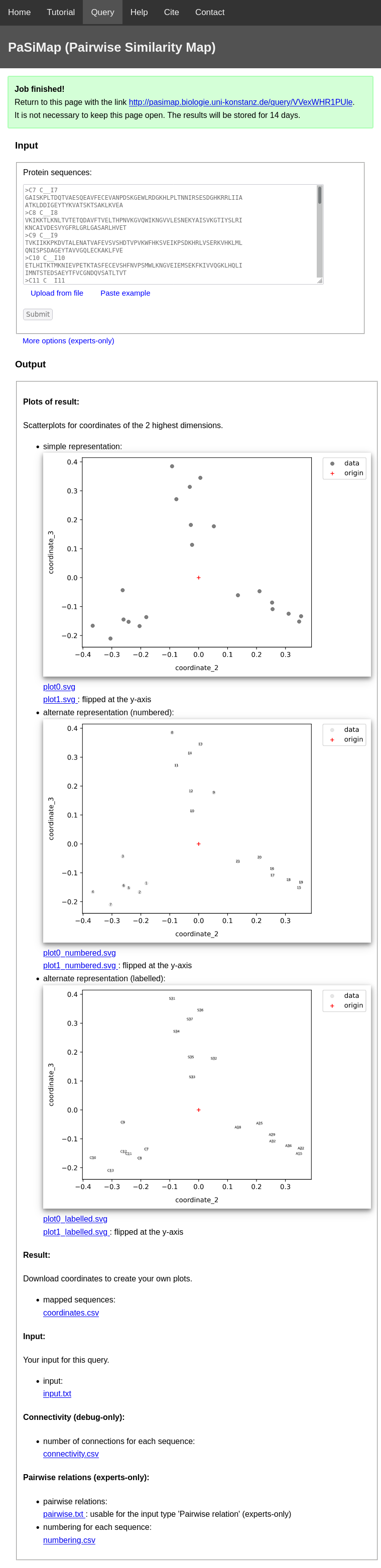
The following output-files can be downloaded:
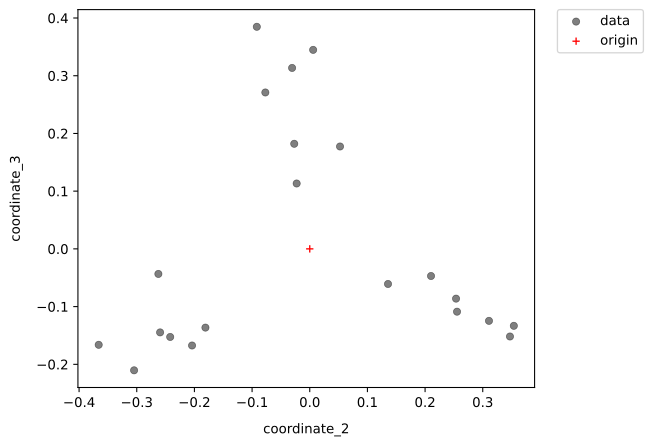
- Plots of result: For your convenience, PaSiMap plots the coordinates for the two highest dimensions of the result.
- simple representation: Each datapoint represents a sequence from your input.
- alternate representation (numbered): The number of the according sequence is plotted on top of each datapoint. The numbering of the sequences can be found in the file 'coordinates.csv'.
- alternate representation (labelled): The first word (separated by
- Result: The file 'coordinates.csv' is the main result of PaSiMap. It contains the coordinates of the mapped protein sequences. These coordinates were used for 'Plots of result'. You can download these coordinates to create your own plots.
- Input: The file 'input.txt' contains the content of the input field for this query. You can download this for easier documentation.
- Connectivity (debug-only): The file 'connectivity.csv' is only relevant, if your job fails because of connectivity-problems. Otherwise, this file can be ignored.
- Pairwise relation (expert-only): These files are only relevant for expert usage and can be ignored.
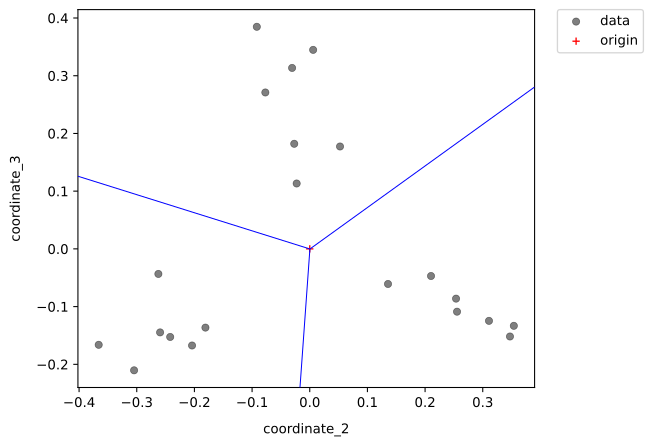
The blue line separates the datapoints into three groups. For more details on the interpretation please refer to Help.
space-character) of the sequence name is plotted on top of each datapoint.
Long labels can result in cluttered plots, so it makes sense to keep the first word of your sequence names short, if you intend to use these plots.
The sequence names correspond to the FASTA-headers of your input protein sequences.
Please note that PaSiMap automatically replaces the special characters ':|,/\' with the character '_'.
Then the first word of each FASTA-header is set as the label.
Afterwards, space-characters of each FASTA-header are also replaced with the character '_', but the label is not affected by this.
Each of these representations can be found in two orientations (flipped at the y-axes). The reason for that is that the absolute values of the coordinates of the mapped protein sequences are not relevant. Instead, the relations between the coordinates are relevant. Therefore, you are free to flip, invert or rotate the coordinates around the origin without changing the interpretation of the result.